
Mohammed El-Kebir
melkebir[AT]illinois[DOT]edu
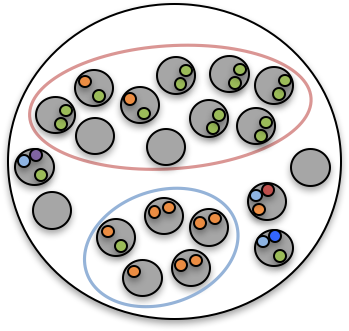
| A tumor is the result of an evolutionary process where somatic mutations are acquired in a population of cells. Somatic mutations occur during the lifetime of an individual and are not inherited. Tumor cells of the same tumor differ in their complement of somatic mutations. This phenomenon is known as intra-tumor heterogeneity and has implications in resistance and treatment. My research focuses on developing algorithms to study the progression of tumors in the context of intra-tumor heterogeneity. Specifically, I have developed algorithms for inferring |
|
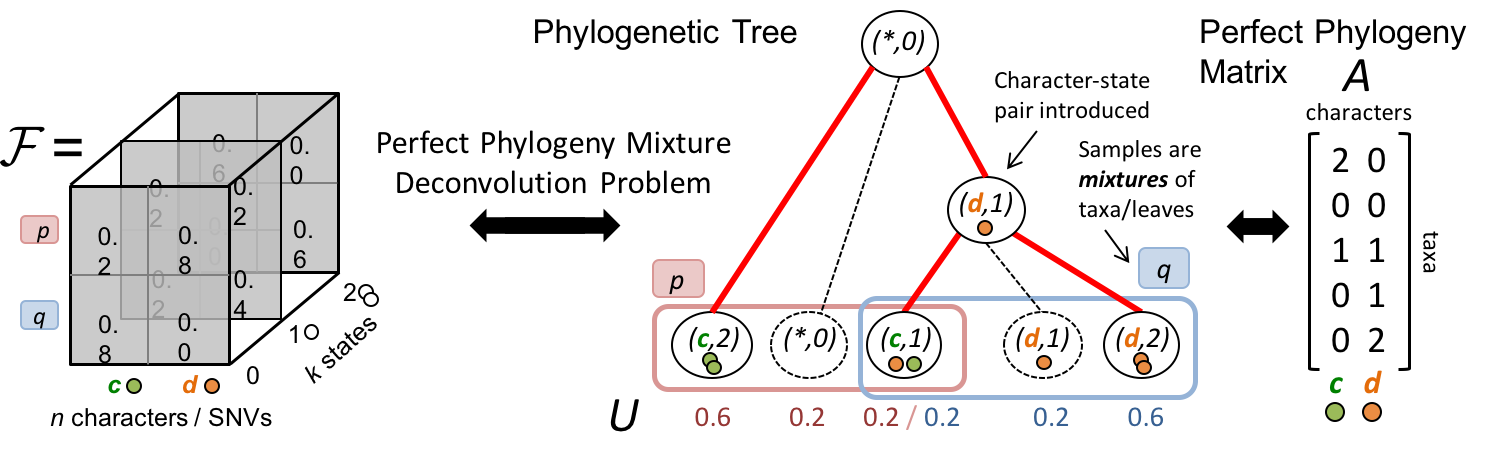
| Most cancer sequencing studies are performed using bulk sequencing technology, where each sample is composed of short reads that originate from the genomes of thousands of cells. In contrast to phylogeny inference for species, where the observations directly correspond to the leaves of an unknown phylogenetic tree, bulk-sequencing samples of a tumor are mixtures of the leaves. Thus, we must simultaneously infer the phylogenetic tree and deconvolve the mixed measurements. |
|
| I introduced the Perfect Phylogeny Mixture Deconvolution Problem (PPMDP), which, given copy number calls and VAFs, asks to infer a multi-state perfect phylogeny such that each state for a character is only introduced once in the tree—i.e. the infinite alleles assumption. This problem can be viewed as $k$ simultaneous constrained nonnegative matrix factorization problems ${F = U A_i}$ that share the matrix $U$ for each state $i \in \{0,\ldots,k-1\}$. |
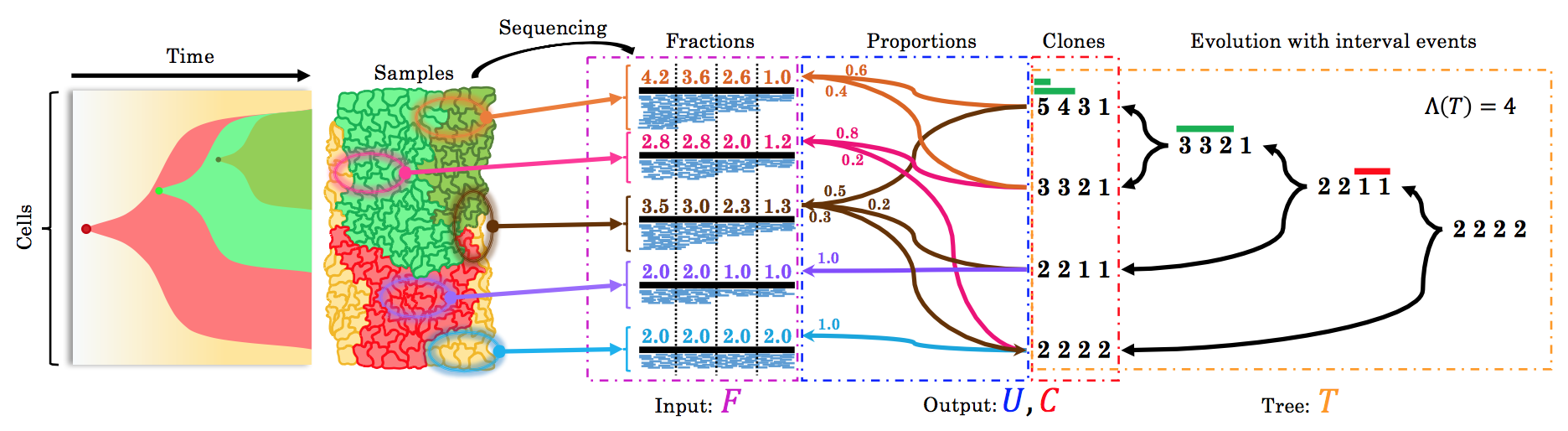
| Current methods are only able to consider single bulk sequencing samples and are thus unable to leverage the additional signal provided by multiple samples. In addition, they do not incorporate an evolutionary model that aims to explain the evolution of the CNAs. I developed a copy-number caller that views samples as mixtures of the leaves of a phylogenetic tree, whose vertices are labeled by a copy-number vector. The mutational events on the edges of this tree correspond to segmental amplifications and deletions of consecutive genomic regions. |
|
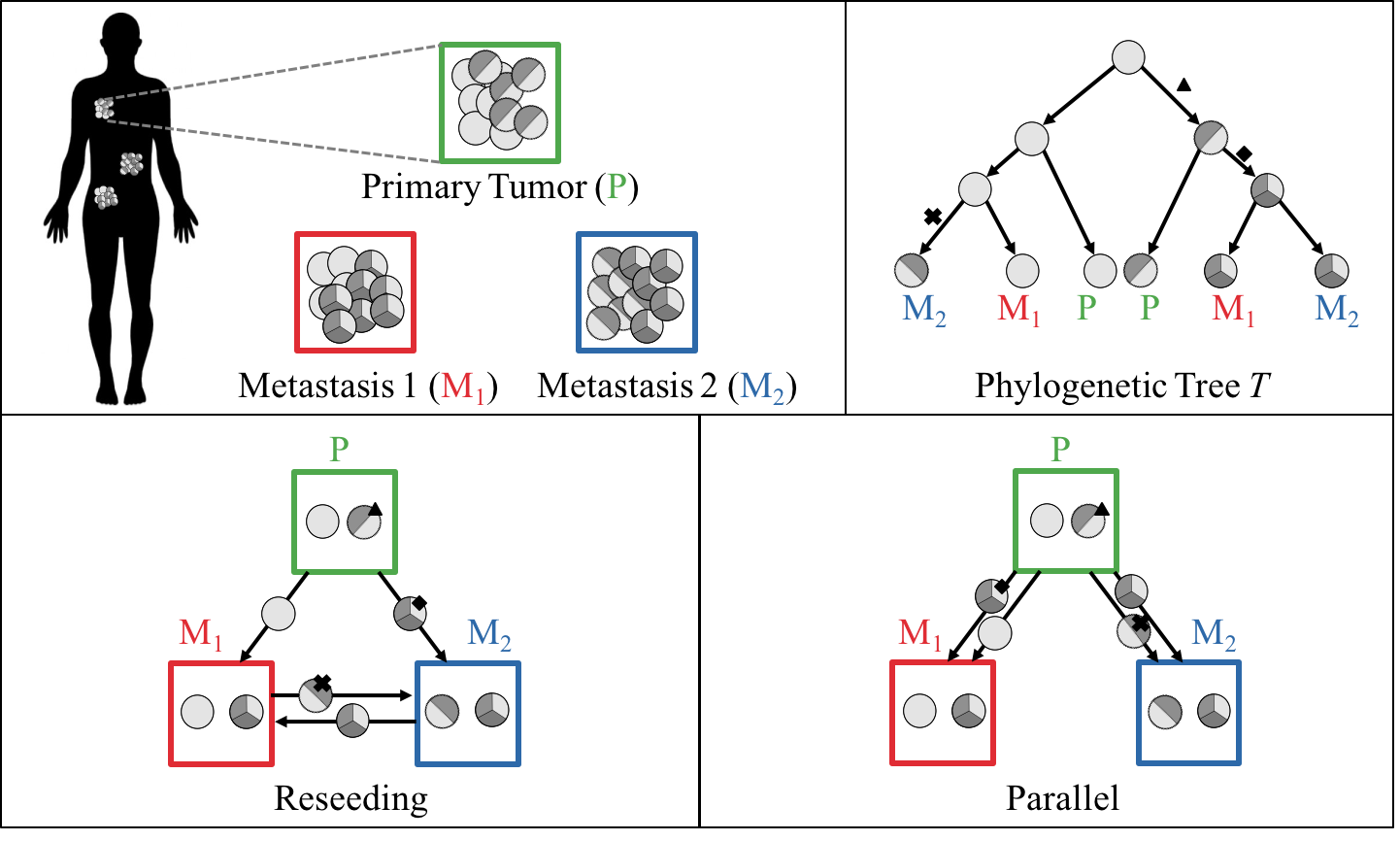
| A tumor cell may also migrate to an anatomical site distant from the primary tumor—this process is called metastasis. The migration process is more complicated than the mutational history of a tumor and cannot always be represented by a tree: groups of cells may migrate together (polyclonal seeding), or the descendant cells of a previous migration may migrate back to the parental site (reseeding). Thus far, the analysis of migration histories has been conducted using manual analysis or ad hoc methods that overlook more parsimonious explanations. |
|
| I introduced a framework that models migration events with a directed multigraph, called the migration graph, and described how to find parsimonious migration patterns. I found that several previously published phylogenetic analyses of metastatic cancers report evolution and migration scenarios that are less biologically plausible than my analyses. |